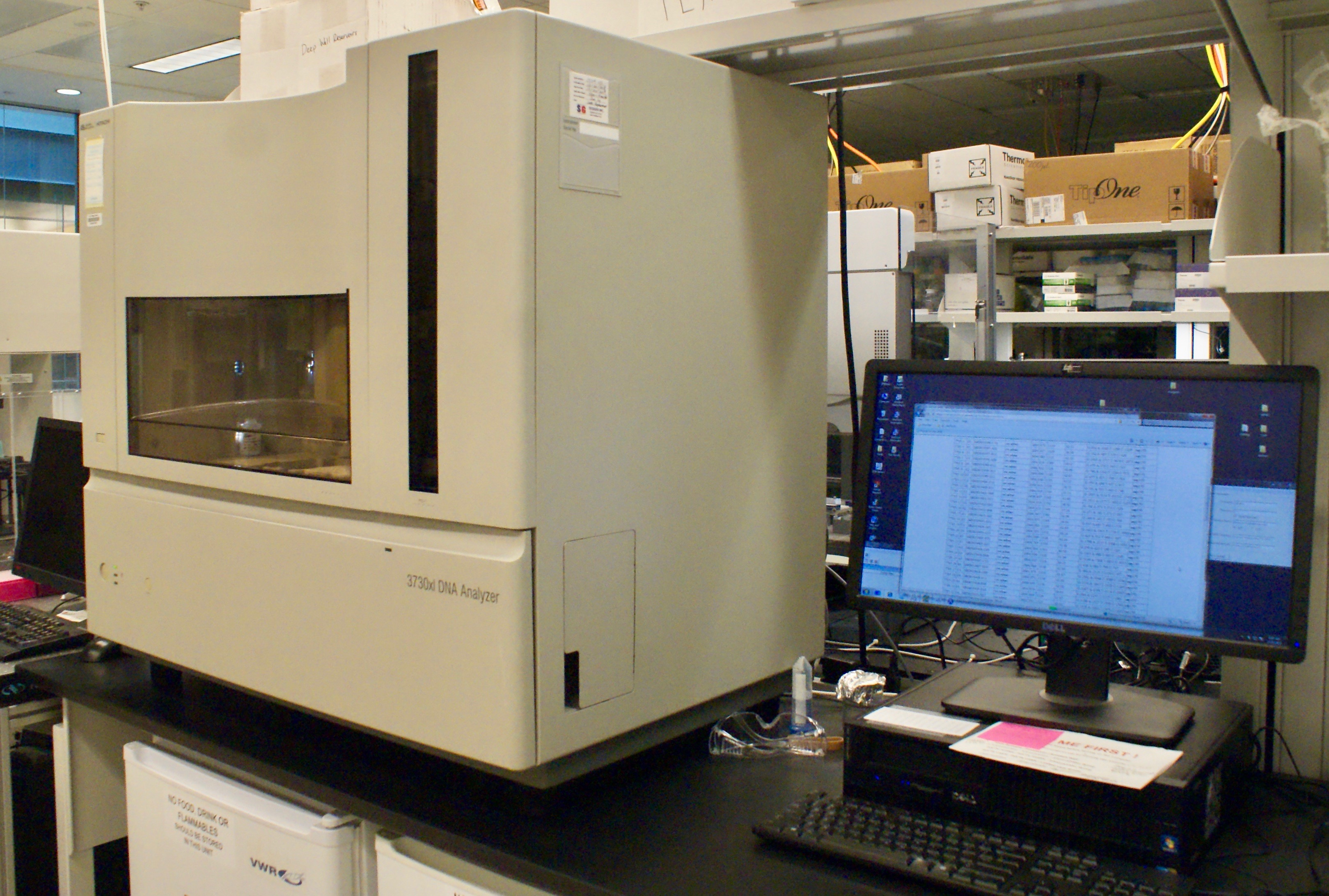
The 3730XL is a 96-Capillary Array DNA sequencer used for DNA fragment analysis and traditional DNA sequencing. Provides quality data at a low cost per sample.
The ABI 3730XL sequencer, from ThermoFisher, is a high-throughput long-read Sanger sequencing instrument. For high quality DNA samples, it can determine about 850bp in a single read, and can read 96 samples simultaneously.
As with any Sanger sequencer, the 3730XL requires samples to be amplified using a single primer and fluorescently-tagged nucleotides. Whenever a tagged nucleotide is incorporated into a new strand, it prevents the polymerase from adding further bases; random incorporation of these tagged nucleotides as compared to untagged nucleotides results in a pool of oligos ranging in length from about 100bp to 1000bp long following PCR cleanup. Capillary electrophoresis on the 3730XL separates these oligos by length and draws them toward an optical system at the end of the capillaries. When the oligos of each consecutive length fluoresce under the laser, the respective intensities for each base’s wavelength are recorded and the location is assigned a base call according to which wavelength has the highest intensity. The resulting output file is called a “trace file” (a portion of which displayed below), and the complete sequence it contains is the compilation of these calls.
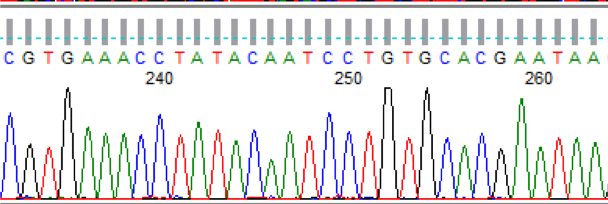
At the Genomics Facility, we manually double-check the automatic base calls determined by the ABI software before returning your data, and rerun samples immediately if reads have poor quality due to excess DNA or dye peaks. For poor quality caused by contamination or insufficient DNA, we will gladly rerun samples if additional clean sample is provided.
Please see below for information on retrieving and interpreting Sanger Sequencing data. This is a dropbox file so if you are unable to open the document please contact genomicshelp@asu.edu and we can send the pdf.
- Sanger sequencing
- FileABI 3730XL manual.pdf7.66 MB
Please see our rates page for detailed pricing information.