Many labs in the Phoenix Metro area use the Regenerative Medicine and Bioimaging Facility to obtain publication quality images. Below is a highlight of some of our more frequent customers.
Tatiana Ugarova’s Lab, ASU School of Life Sciences
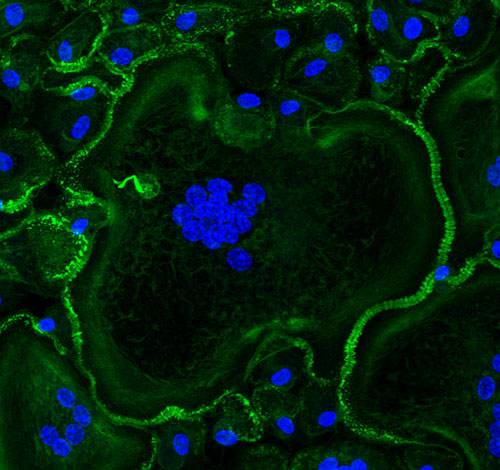
Dr. Ugarova’s lab studies the cellular and molecular mechanisms underlying cell-cell fusion, a process essential for both biology and medicine. In particular, macrophage fusion resulting in the formation of destructive multinucleated giant cells (MGCs) is a hallmark of numerous maladies associated with chronic inflammation, including granulomatous infections, atherosclerosis, amyotrophic lateral sclerosis, the foreign body reaction to implanted biomaterials, cancer and others. We are interested in the role of the actin cytoskeleton, integrins and other transmembrane receptors that mediate this process. We have recently revealed that macrophage fusion is initiated by an actin-based protrusion originating from a region enriched in podosomes. Instrumental to this discovery were phase-contrast and confocal video microscopy available in the Regenerative Medicine and Bioimaging Facility. Using these and other imaging methods, we have recently identified previously unrecognized actin-based structures formed in mature MGCs that we believe promote undesirable survival of these cells on the surface of implanted biomaterials. To better understand the mechanisms involved in macrophage fusion we plan to expand these studies using living specimens that will be analyzed by multiphoton microscopy and confocal microscopy equipped with the light sheet module.
-
Faust, J.J., Christenson, C., Doudrick, K., Ros, R. Ugarova, T.P. (2017) Development of fusogenic glass surfaces that impart spatiotemporal control over macrophage fusion: Direct visualization of multinucleated giant cell formation, Biomaterials, 128: 160-171
- Faust, J.J., Christenson, C., Doudrick, K., Heddleston, J., Chew, T-L., Lampe, M., Balabiyev, A., Ros, R., Ugarova, T.P. (2018) Fabricating optical-quality glass surfaces to study macrophage fusion. Journal of Visualized Experiments, 133: 56866
- Lishko, V.K., Yakubenko, V.P., Ugarova, T.P., Podolnikova, N.P. (2018) Leukocyte Integrin Mac-1 (CD11b/CD18, αMβ2, CR3) Acts as a Functional Receptor for Platelet Factor 4, Journal of Biological Chemistry, 293, 6869-6882.
- Cui, K., Ardell, C.L., Podolnikova, N.P., and Yakubenko, V.P. (2018) Distinct Migratory Properties of M1, M2, and Resident Macrophages Are Regulated by αDβ2 and αMβ2 Integrin-Mediated Adhesion. Frontiers in Immunology, 2650 (9), 1-14.
- Faust, J.J., Balabiyev, A., Heddleston, J.M., Podolnikova, N.P., Baluch, D.P., Chew, T.L., and Ugarova, T.P. (2019) An actin-based protrusion originating from a podosome-enriched region initiates macrophage fusion. Molecular Biology of the Cell, In Review.
- Podolnikova, N.P., Hlavackova, M., Wu, Y., Yakubenko, V.P., Faust, J.J., Balabiyev, A., Ugarova, T.P. (2019) Interaction between the integrin Mac-1 (CD11/CD18) and signal regulatory protein α (SIRPα) mediates fusion in heterologous cells. Journal of Biological Chemistry, PMID: 30910815
Mike Kruer’s Lab, Barrow Neurological Institute at Phoenix Children's Hospital
The Kruer lab is within the Barrow Neurological Institute at Phoenix Children's hospital. Our research focuses on understanding the contributions rare genetic variants play in neurologically based movement disorders such as cerebral palsy. We use a multi-pronged approach to understand genetic causes of idiopathic (no obvious environmental etiology) cerebral palsy including whole exome trio studies followed by functional validation of discovered genes in Saccharomyces, Drosophilia, and patient derived iPSC neuron model systems. We have identified many novel gene variants, which appear to lead to a cerebral palsy phenotype. Many of these variants fall in genes associated with cytoskeletal regulation thus fluorescent microscopy lends itself nicely to in vitro cellular morphometry.
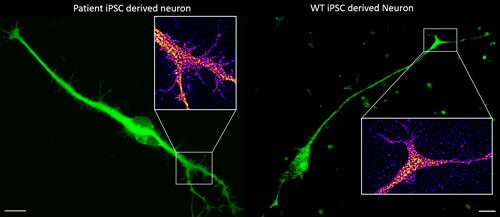
The patient neuron (left) harbors a mutant variant of a gene suspected to play a role in the regulation of cellular morphology and neurite outgrowth. Closely comparing the periphery of the Map2 labeled patient neuron vs our wild-type (right) neuron (insets) supports our preliminary enzymatic molecular data and yeast functional studies suggesting that our identified mutation does alter the protein's functionality. Images acquired on the Leica SP8 at the Regenerative Medicine and Bioimaging Facility. Scale bar = 10 microns.
Stuart Newfeld’s Lab, ASU School of Life Sciences
CORL proteins (known as SKOR in mice, Fussel in humans and fussel in Flybase) are a family of CNS specific proteins related to Sno/Ski oncogenes. Their developmental and adult roles are largely unknown. A Drosophila CORL (dCORL) reporter gene is expressed in all Drosophila insulin-like peptide 2 (dILP2) neurons of the pars intercerebralis (PI) of the larval and adult brain. The transcription factor Drifter is also expressed in the PI in a subset of dCORL and dILP2 expressing neurons and in several non-dILP2 neurons. dCORL mutant virgin adult brains are missing all dILP2 neurons that do not also express Drifter. This phenotype is also seen when expressing dCORL-RNAi in neurosecretory cells of the PI. dCORL mutant virgin adults of both sexes have a significantly shorter lifespan than their parental strain. This longevity defect is completely reversed by mating (lifespan increases over 50% for males and females). Analyses of dCORL mutant mated adult brains revealed a complete rescue of dILP2 neurons without Drifter. Taken together, the data suggest that dCORL participates in a neural network connecting the insulin-signaling pathway, longevity and mating. The conserved sequence and CNS specificity of all CORL proteins imply that this network may be operating in mammals.
- Tran, N.L., Goldsmith, S.L. Dimitriadou, A., Takaesu, N.T., Consoulas, C. & Newfeld S.J (2018b) CORL expression and function in insulin producing neurons reversibly influences adult longevity in Drosophila. G3: Genes, Genomes, Genetics, 8:2979-2990.
- Tran, N., Takaesu, N., Cornell, E. & Newfeld S.J. (2018a) CORL expression in the Drosophila central nervous system is regulated by stage specific interactions of intertwined activators and repressors. G3: Genes, Genomes, Genetics, 8:2527-2536.
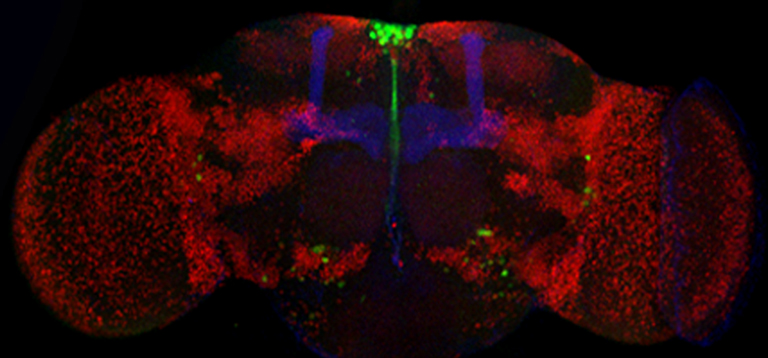
The Drosophila CORL protein is expressed in all insulin producing neurons of the adult brain. Whole mount confocal view of a one day old adult female brain. This brain was histologically stained for three proteins: dCORL (green), Fas2 (blue – handlebar shape that marks a specific neuropil providing orientation) and Toy (red – identifies a vareity of specific neuropil by marking the nucleus of many brain neurons). dCORL is strongly expressed in the dorsal midline region called the pars intercerebralis. This is a well-known neurosecretory region of the brain provides insulin.

